A brief visualization of R's distribution functions, focusing on the normal distribution
The intent of this post is to provide a visual intuition of the various distribution functions in R, using the normal distribution as an example.
When I first started playing with the distribution functions in R I would always get confused as to what the different name combinations meant. For example, the differences between rnorm(), dnorm(), dt(), rbinom() were all pretty mysterious to me. Now I understand these names are composed of two parts:
- Prefix: An indication of the “map” being made (more on that in a moment)
- Suffix: The distribution of interest.
In terms of understanding the suffix, R comes pre-loaded with a bunch of typical distributions, such as (note the use of "*" as a place-holder for the prefixes of R’s distribution function names):
*norm: Normal*t: Student-t*binom: Binomial
That was kind of an “A ha!” moment, and seems pretty obvious now. I should also mention a complete list of R’s built-in distributions can be found here. There is also a really, really useful CRAN Task View focused specifically on distributions.
What was less obvious to me was what the various prefixes on the distribution names meant. Typically, these prefixes and their meanings are ("*" simply indicates there is usually a suffix associated with these functions):
d*: Returns a density (point probability; likelihood) value, given a quantile and some set of distribution parameters.p*: Returns a cumulative probability value, given a quantile and some set of distribution parameters.q*: Returns a quantile, given a cumulative probability value and some set of distribution parameters.r*: Returns a random number (quantile), the value of which is determined by a distribution described by some set of parameters.
The above meanings were not obvious to me from the help files that came with R, nor were the mathematical descriptions therein. Instead what I found to be useful were visual representations of these different relationships, which helped me better conceptualize the different “mappings” associated with the prefixes highlighted above.
Thus, the point of this post is to provide myself (and hopefully others) with a visual intuition of the various distribution functions in R, focusing on the normal distribution as an example.
Libraries
We’ll use a couple of libraries to help illustrate the relationships. patchwork is a great library for putting together different ggplot2 outputs that would be otherwise difficult to combine. paletteer is my favorite color package, as it aggregates a number of other color packages, making available a wide selection of pretty color combinations. And then the tidyverse loads ggplot2, as well as other helpful packages such as dplyr and tidyr.
library(patchwork)
library(paletteer)
library(tidyverse)Making the data
To explore the relationship between the different normal distribution functions I’ll use a standard normal distribution, which is centered on 0 (i.e., the mean is 0) and has a standard deviation of 1.
#Data and parameters
z_scores <- seq(-4, 4, by = 0.01)
mu <- 0
sd <- 1
#Functions
##Using `dnorm` and `pnorm` to setup the "skeleton" of related plots.
normal_dists <- list(`dnorm()` = ~ dnorm(., mu, sd),
`rnorm()` = ~ dnorm(., mu, sd),
`pnorm()` = ~ pnorm(., mu, sd),
`qnorm()` = ~ pnorm(., mu, sd))
##Apply functions to data and parameter combinations
df <- tibble(z_scores, mu, sd) %>%
mutate_at(.vars = vars(z_scores), .funs = normal_dists) %>%
#"Lengthen" the data
pivot_longer(cols = -c(z_scores, mu, sd), names_to = "func",
values_to = "prob") %>%
#Categorize based on shape of distribution -- need to split up the dataframe
# for plotting later.
mutate(distribution = ifelse(func == "pnorm()" | func == "qnorm()",
"Cumulative probability", "Probability density"))
##Split up the data into different pieces that can then be added to a plot.
###Probabilitiy density distrubitions
df_pdf <- df %>%
filter(distribution == "Probability density") %>%
rename(`Probabilitiy density` = prob)
###Cumulative density distributions
df_cdf <- df %>%
filter(distribution == "Cumulative probability") %>%
rename(`Cumulative probability` = prob)
###dnorm segments
#Need to make lines that represent examples of how values are mapped -- there
# is probably a better way to do this, but quick and dirty is fine for now.
df_dnorm <- tibble(z_start.line_1 = c(-1.5, -0.75, 0.5),
pd_start.line_1 = 0) %>%
mutate(z_end.line_1 = z_start.line_1,
pd_end.line_1 = dnorm(z_end.line_1, mu, sd),
z_start.line_2 = z_end.line_1,
pd_start.line_2 = pd_end.line_1,
z_end.line_2 = min(z_scores),
pd_end.line_2 = pd_start.line_2,
id = 1:n()) %>%
pivot_longer(-id) %>%
separate(name, into = c("source", "line"), sep = "\\.") %>%
pivot_wider(id_cols = c(id, line), names_from = source) %>%
mutate(func = "dnorm()",
size = ifelse(line == "line_1", 0, 0.03))
###rnorm segments
#Make it reproducible
set.seed(20200209)
df_rnorm <- tibble(z_start = rnorm(10, mu, sd)) %>%
mutate(pd_start = dnorm(z_start, mu, sd),
z_end = z_start,
pd_end = 0,
func = "rnorm()")
###pnorm segments
df_pnorm <- tibble(z_start.line_1 = c(-1.5, -0.75, 0.5),
pd_start.line_1 = 0) %>%
mutate(z_end.line_1 = z_start.line_1,
pd_end.line_1 = pnorm(z_end.line_1, mu, sd),
z_start.line_2 = z_end.line_1,
pd_start.line_2 = pd_end.line_1,
z_end.line_2 = min(z_scores),
pd_end.line_2 = pd_start.line_2,
id = 1:n()) %>%
pivot_longer(-id) %>%
separate(name, into = c("source", "line"), sep = "\\.") %>%
pivot_wider(id_cols = c(id, line), names_from = source) %>%
mutate(func = "pnorm()",
size = ifelse(line == "line_1", 0, 0.03))
###qnorm segments
df_qnorm <- tibble(z_start.line_1 = min(z_scores),
pd_start.line_1 = c(0.1, 0.45, 0.85)) %>%
mutate(z_end.line_1 = qnorm(pd_start.line_1),
pd_end.line_1 = pd_start.line_1,
z_start.line_2 = z_end.line_1,
pd_start.line_2 = pd_end.line_1,
z_end.line_2 = z_end.line_1,
pd_end.line_2 = 0,
id = 1:n()) %>%
pivot_longer(-id) %>%
separate(name, into = c("source", "line"), sep = "\\.") %>%
pivot_wider(id_cols = c(id, line), names_from = source) %>%
mutate(func = "qnorm()",
size = ifelse(line == "line_1", 0, 0.03))Plotting the relationships
Now that the data has been made, we can put it all together using ggplot2 and patchwork.
#Plot the data
# Note: I can't combine the data in a normal facet layout, due to differences
# in axis labels, so I'm using the patchwork library to bring everything
# together after the fact.
##Color palette
cp <- paletteer_d("ggsci::default_locuszoom", 4, )
names(cp) <- c("dnorm()", "rnorm()", "pnorm()", "qnorm()")
##Probabilitiy density
p_pdf <- df_pdf %>%
ggplot(aes(z_scores, `Probabilitiy density`)) +
geom_segment(data = df_dnorm,
aes(z_start, pd_start, xend = z_end, yend = pd_end),
arrow = arrow(length = unit(df_dnorm$size, "npc"), type = "closed"),
size = 0.8, color = cp["dnorm()"]) +
geom_segment(data = df_rnorm,
aes(z_start, pd_start, xend = z_end, yend = pd_end),
arrow = arrow(length = unit(0.03, "npc"), type = "closed"),
size = 0.8, color = cp["rnorm()"]) +
geom_line(size = 0.6) +
facet_wrap(~ func, nrow = 1) +
theme_bw() +
theme(panel.grid = element_blank(),
axis.title.x = element_blank(),
strip.background = element_blank(),
text = element_text(family = "serif", size = 14)) +
scale_y_continuous(expand = expand_scale(c(0, 0.05))) +
scale_x_continuous(expand = c(0.01, 0))
##Cumulative probability
p_cdf <- df_cdf %>%
ggplot(aes(z_scores, `Cumulative probability`)) +
geom_hline(yintercept = 0, color = "grey") +
geom_segment(data = df_pnorm,
aes(z_start, pd_start, xend = z_end, yend = pd_end),
arrow = arrow(length = unit(df_dnorm$size, "npc"), type = "closed"),
size = 0.8, color = cp["pnorm()"]) +
geom_segment(data = df_qnorm,
aes(z_start, pd_start, xend = z_end, yend = pd_end),
arrow = arrow(length = unit(df_qnorm$size, "npc"), type = "closed"),
size = 0.8, color = cp["qnorm()"]) +
geom_line(size = 0.6) +
facet_wrap(~ func, nrow = 1) +
labs(x = "z-score/quantiles") +
theme_bw() +
theme(panel.grid = element_blank(),
strip.background = element_blank(),
text = element_text(family = "serif", size = 14)) +
scale_x_continuous(expand = c(0.01, 0))
##Combine the plots
p_pdf + p_cdf + plot_layout(ncol = 1)
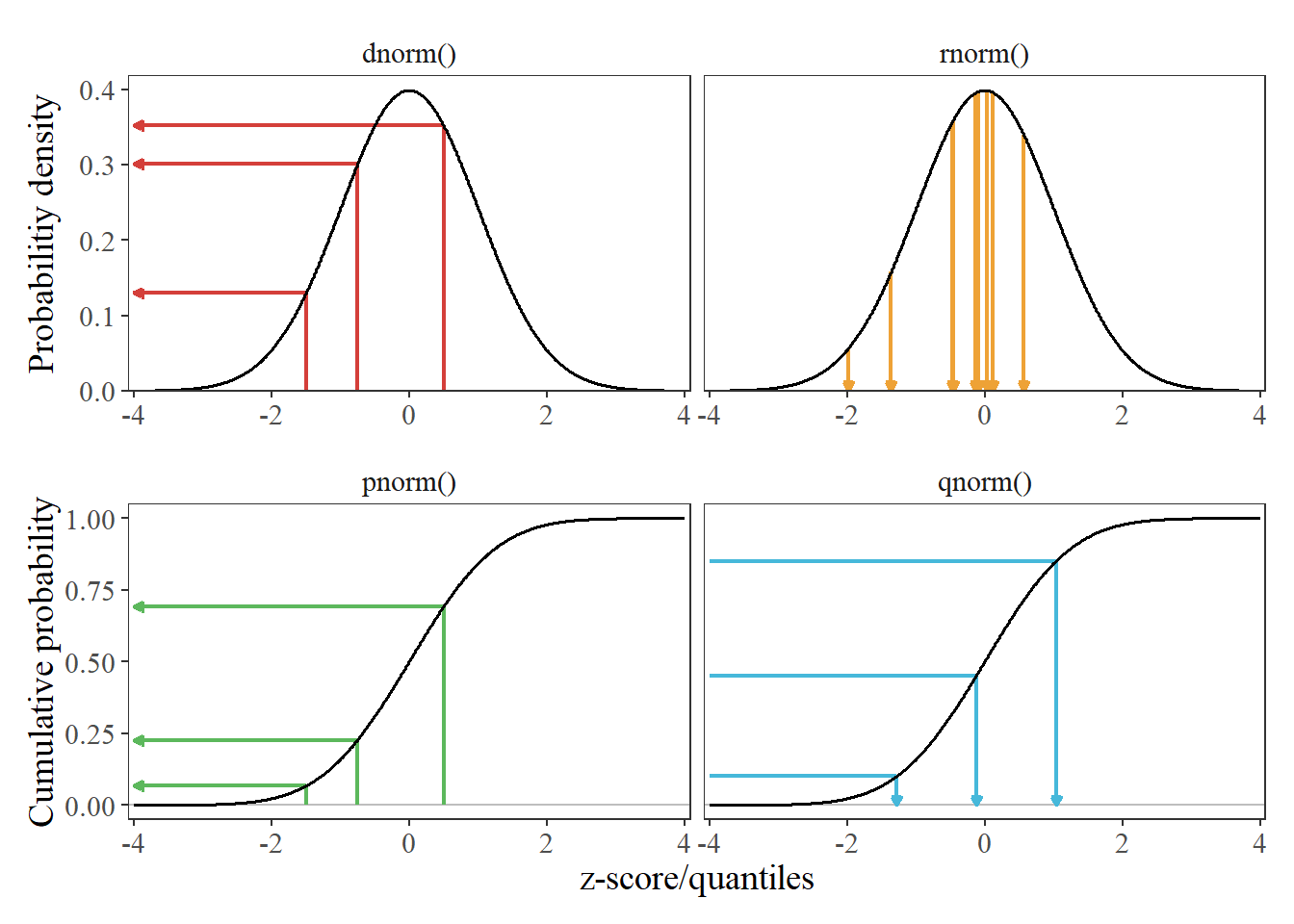
Figure 1: Examples comparing the mappings between different normal distribution functions, given a mean of 0 and standard deviation of 1. Because a standard normal distribution is being used, the terms ‘z-score’ and ‘quantile’ are used interchangeably. Arrows are intended to provide a sense of direction for the examples, in terms of what data are input and output from a given function. dnorm() maps z-scores to their respective density values (point probabilities). rnorm() randomly draws quantiles, weighted by their probability – hence why the most of the 10 random draws are clustered near the peak of the distribution. pnorm() maps a z-score to it’s cumulative probability. qnorm() maps a cumulative probability to a quantile.
And there we have it! A simple visualization to provide a little bit more intuition into R’s distribution functions.